Statistical physics & evolutionary biology to understand living matter, from allostery and catalysis in proteins or artificial systems to gene regulation in bacterial genomes and adaptation in populations.
Natural selection is fundamental to biology but foreign to physics. Why is it so? Can we design a simple physical system that undergoes evolution by natural selection?
Enzymes are unrivalled catalysts. Are they following special design principles? Can we make comparable artificial catalysts with programmable matter?

Evolution is a matter of information processing. How to formalize this intuition and rationalize the diversity of transmission mechanisms, from Lamarckism to sex?

In bacteria, genetic context is more evolutionarily conserved than transcription factors. How does it regulate gene expression? How to predict its impact?

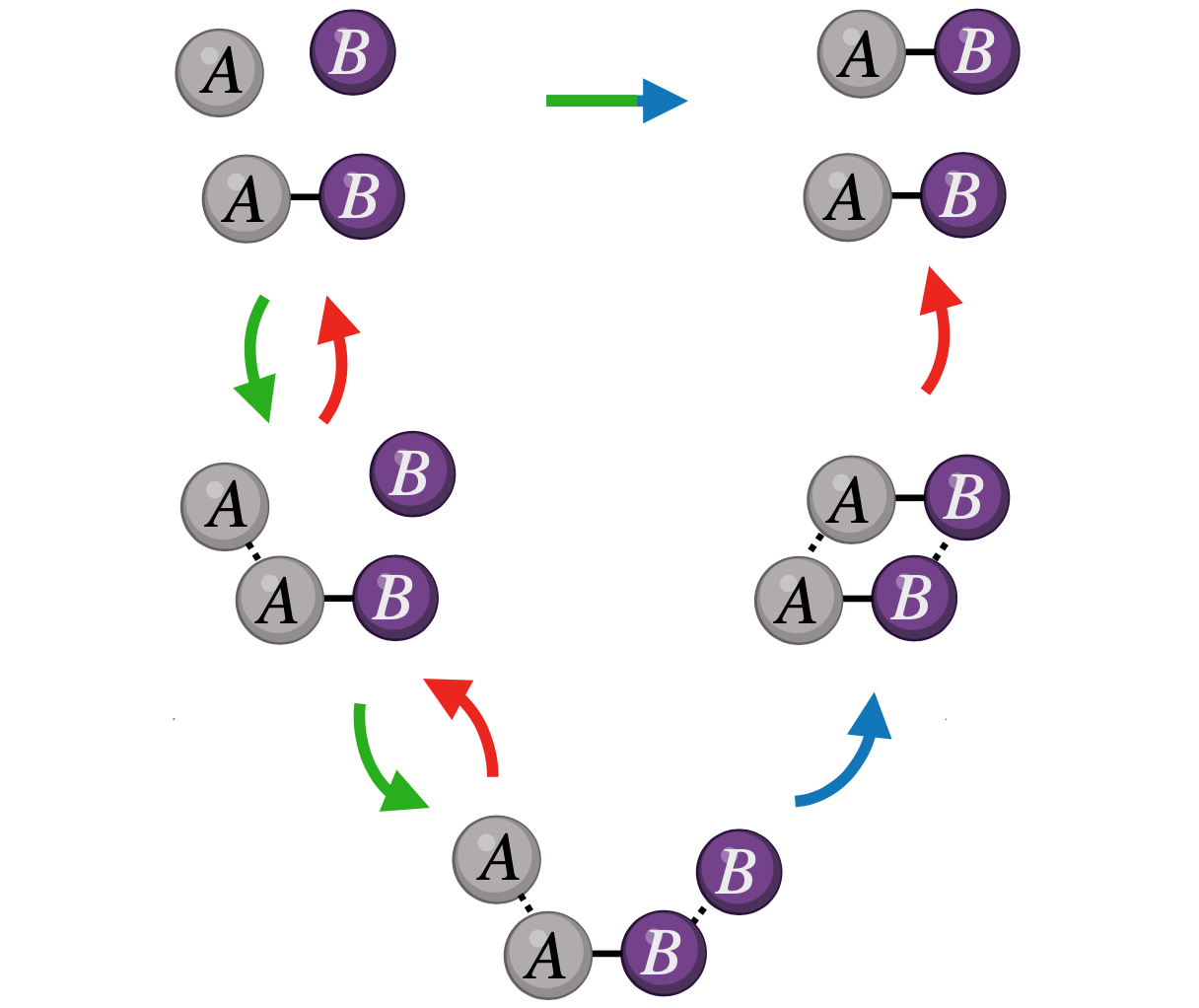
Allostery is the regulation of protein activities by perturbations away from their active site. How does it work? Why is it so widespread?
The basic elements of evolution – selection/amplification/mutations – can be reproduced in the lab. But how to navigate the gigantic sequence space?

