Theory of allostery
Allostery is the regulation of protein activities by perturbations away from their active site. How does it work? Why is it so widespread?
Evolving allostery in a physical model
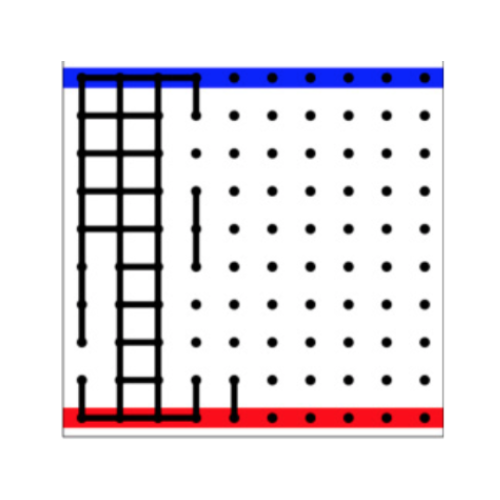
Allostery is intriguing from both a physical and evolutionary perspective. Physically, how do long-range effects arise from local interactions? Evolutionarily, how do they arise from past selective pressures? We have proposed a first model that combines physical and evolutionary constraints and used it to show how an allosteric channel connecting two distant sites can evolve, the shape of which depends on evolutionary history.
Reference: M. Hemery, O. Rivoire (2015). Evolution of sparsity and modularity in a model of protein allostery.
Origins of latent allostery
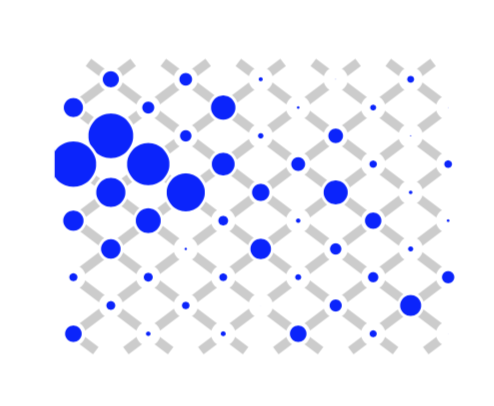
Many, if not all, proteins exhibit long-range effects, even in the absence of allosteric regulation. This “latent allostery” is considered the basis on which allosteric regulation can evolve, but how to explain its origin? We proposed a scenario in which selection for discriminative binding requires a conformational switch that involves distant sites and showed how this scenario can generate the different coevolutionary patterns observed in protein alignments.
Reference: O. Rivoire (2019). Parsimonious evolutionary scenario for the origin of allostery and coevolution patterns in proteins.
Single versus multi-state allostery
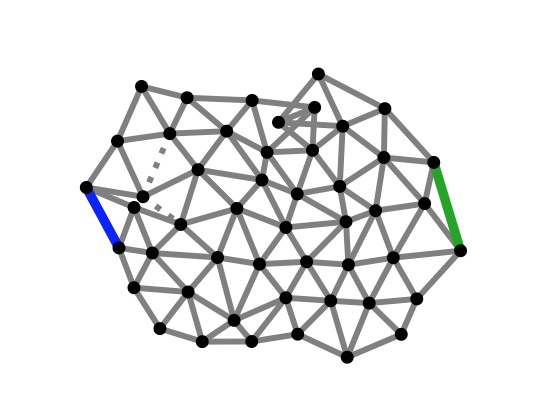
Two phenomenological models of allostery are often contrasted: an induced-fit model (aka KNF model) and a two-state model (aka MWC model). When does evolution “choose” one or the other? We developed a model to answer this question and found that when the two mechanisms can both evolve, the two-state mechanism is favored and provide a larger allosteric effect.
Reference: E. Rouviere, R. Ranganathan, O. Rivoire (2021). On the emergence of single versus multi-state allostery.
